Internal simulation capability include energy minimization and molecular dynamcs.
Molecular Mechanics
Two different algorithms, the conjugate gradient and variable metric methods, are implemented in Direct Force Field for energy minimization. The conjugate gradient method is based on the Polak-Ribiere method, which is similar to the Fletcher-Reeve method. The variable metric method is also called quasi-Newton method. Both methods involve calculating the derivatives of the potential energy.
Restrained energy minimization can be performed to explore energy profiles. This is done by adding energy terms on selected internal coordinates to the total energy function. The restraint function is a harmonic function in which the reference value and force constant can be specified. The restraint energy value is subtracted from the total energy in the results reported.
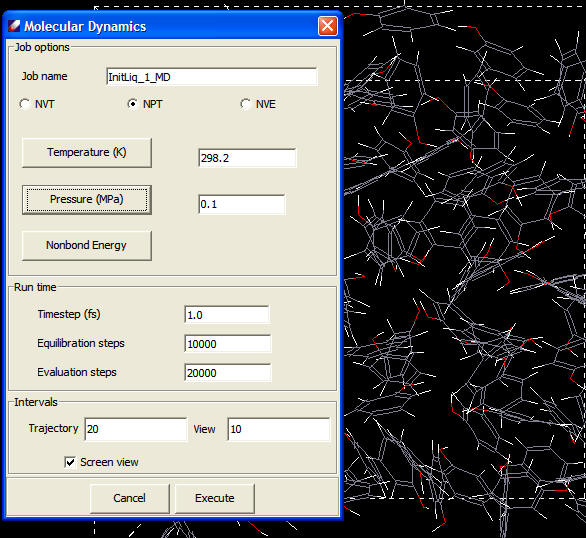
Molecular Dynamics
The Verlet velocity algorithm is implemented in Direct Force Field. The available ensembles are:
- NVE (microcanonical)
- NVT (canonical)
- NPT (isothermal-isobaric)
The temperature can be modularized using two methods:
- Direct Velocity Scaling
- Andersen Stochastic Method
Pressure changes can be accomplished by changing the coordinates of the particles and the size of the unit cell in periodic boundary conditions. Berendsen's method couples the system to a pressure "bath" to maintain the pressure at a certain target. The following screenshot shows the MD job dialog.